|
The Most Effective Deconvolution Package Available
Huygens makes intelligent use of a microscope's inherent imaging characteristics, which produces higher image resolution and lower noise levels than other deconvolution systems. Depending on the type of image you have and the quality you need, the Huygens system offers three deconvolution options:
• Maximum Likelihood Estimation (MLE)
• Iterative Constrained Tikhonov-Miller restoration
• Non-iterative Tikhonov-Miller algorithm.
Huygens analyzes your image and makes informed, image-specific recommendations for optimum parameter settings. It offers specific algorithms for wide-field, confocal, 2-photon imaging, scanning disk and 4Pi microscopy.
In addition to advanced restoration algorithms, Huygens:
• Supports 2D and 3D images acquired over time
• Leverages multi-processor systems including those with more than 100 processors
• Stores tasks in interactive mode as Tool Command Language (TCL) commands for automated deconvolution in batch mode
• Supports 64-bit Irix® operating system
• Enables parallel processing of sub tasks
• Allows the use of more than 4GB of RAM to streamline large image processing
Huygens is available in two versions: Huygens Professional—a full-featured software package for the Irix and Linux operating systems and Huygens Essential—based on the same deconvolution engine as Huygens Professional and compatible with Microsoft Windows® 2000™, Irix and Linux.
The developer of the Huygens system, Dr. H. T. M. van der Voort, is one of the pioneers of image restoration technology. Dr. van der Voort has invented many of the most recognized and well-documented deconvolution techniques. The algorithms that form the basis of the HUYGENS system have been validated and field tested for many years. More than a dozen papers in scientific journals such as the Journal of Microscopy describe the functions of the Huygens system.
View the deconvolution image gallery.
|
 |
 |
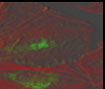 |
 Before using Huygen's deconvolution. Before using Huygen's deconvolution. |
 |
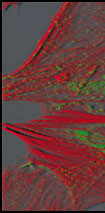 |
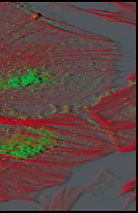 |
 |

After deconvolution using (number) of MLE passes.
Dr. Cecile Gauthier, CRBM, UPR, 1086 CNRS, Montepellier, CRBM Integrated Imaging Facility.
View the deconvolution image gallery.
|
|
|
 |

